Biosynthesis & Bioengineering of Natural Products
Our group studies the role and mechanism of enzymes and other proteins involved in biosynthetic pathways. To this end, the proteins of interest are first produced in a bacterial or fungal heterologous hosts. The proteins are then purified using chromatographic methods such as immobilized metal affinity chromatography and size-exclusion chromatography. The purified proteins can then be used to investigate the biosynthetic pathway, enzyme activity, ligand binding, protein-protein interactions and protein structure.

In order to better understand the function of enzymes on a molecular level, our group utilizes protein X-ray crystallography to determine their structure. First, hundreds to thousands of different conditions are screened to identify the ones in which the protein of interest forms crystals. The growth of the crystals is then optimized, and the crystals are targeted with high-intensity X-rays, so that diffraction patterns can be collected. From these data, a model of the protein can be generated.
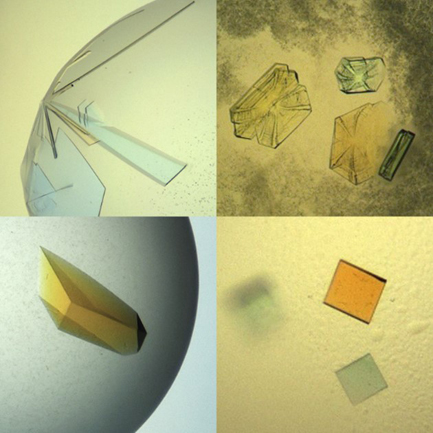
Protein crystals grown in various conditions
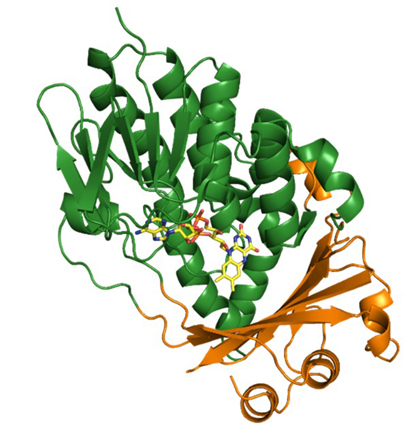
Structure of a biosynthetic enzyme with bound FAD cofactor
Below, a few selected projects are outlined:
- Bacterial aromatic polyketides are a class of natural products with, e.g., antibacterial, antiviral or anticancer properties. The biosynthesis of these compounds starts with the minimal polyketide synthase followed by the action of cyclases and aromatases to yield the initial polycyclic polyketide. These intermediary products are typically further processed to the final natural product by various pathway-specific tailoring enzymes, e.g. oxidases or transferases, that structurally and therefore also functionally diversify these compounds. These elaborate processing steps can be harnessed to create novel “unnatural” natural products by combining various tailoring enzymes (from different pathways) for example in in vitro reactions.

Purified FAD-dependent biosynthetic enzyme
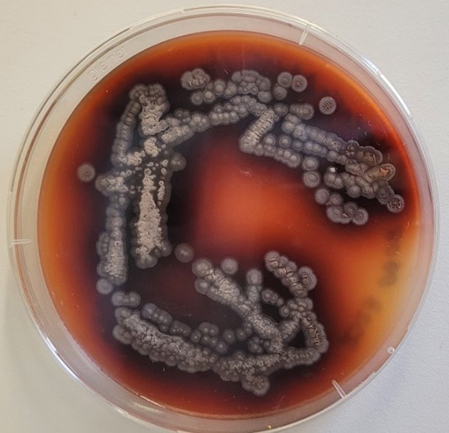
Streptomyces producing natural product on solid medium
- Tropone natural products are a structurally unique class of secondary metabolites with diverse biological activities. However, their biosynthetic pathways and ecological roles remain poorly understood. Using a combination of metabolomics and genetic methods, we aim to investigate the microbial production of tropones and the regulatory factors influencing their biosynthesis. Additionally, the underlying enzymology of these biosyntheses is scrutinized using biochemical and biophysical methods. We explore microbial interactions as potential modulators of natural product biosynthesis, hypothesizing that symbiotic or competitive relationships impact the activation of cryptic biosynthetic gene clusters.
Lab Impressions

Cultivation of antibiotic-producing bacteria.

A co-cultivation chamber to study chemical interactions of physically separated organisms.

Bacteria (center) produce compounds that inhibit the growth of a fungus.
- Secondary metabolism in plants: Recent major advances in sequencing techniques pave the way to facilitate the study on plant enzymes involved in pharmacologically interesting biosynthetic pathways. Finding such key enzymes is complicated in plants due to, e.g., splicing and scattering of the encoding genes across the genome, although occasional clustering of enzymes involved in a certain biosynthetic pathway can be observed. Still, several approaches are often required to find the correct enzyme candidate, e.g., transcriptomic and genomic data, native protein purification and/or pull-down assays, which can provide a selection of candidates for further in vitro testing.
The availability and stability of enzyme substrates is often also a problem, which has to be addressed, e.g., by natural product extraction, chemical synthesis or engineering of a proper yeast or plant host to access the desired substrate. If conversion assays with the enzymes heterologously expressed in E. coli do not result in successful product formation, assays need to be reconstituted in yeast expression systems or via Agrobacterium-mediated transformation of Nicotiana species.
Plant material which is used in our lab to study plant biosynthetic enzymes.


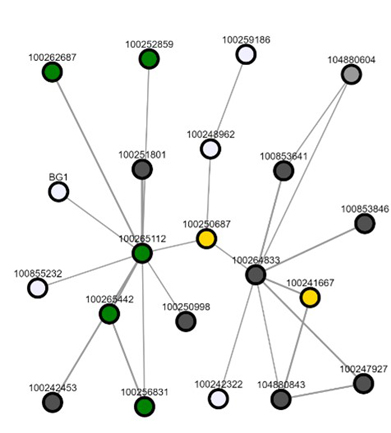
Co-expression network of transcriptomic data used to identify candidate enzymes for in vitro testing.
